Fitting single subject data using Bayesian estimation#
Author: Nicolas Legrand nicolas.legrand@cfin.au.dk
%%capture
import sys
if 'google.colab' in sys.modules:
! pip install metadpy
import arviz as az
import numpy as np
from metadpy.bayesian import hmetad
From response-signal arrays#
# Create responses data
nR_S1 = np.array([52, 32, 35, 37, 26, 12, 4, 2])
nR_S2 = np.array([2, 5, 15, 22, 33, 38, 40, 45])
This function will return two variable. The first one is a pymc model variable
model, traces = hmetad(nR_S1=nR_S1, nR_S2=nR_S2)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Sequential sampling (4 chains in 1 job)
NUTS: [c1, d1, meta_d, cS1_hn, cS2_hn]
100.00% [2000/2000 00:18<00:00 Sampling chain 0, 0 divergences]
100.00% [2000/2000 00:18<00:00 Sampling chain 1, 0 divergences]
100.00% [2000/2000 00:17<00:00 Sampling chain 2, 0 divergences]
100.00% [2000/2000 00:18<00:00 Sampling chain 3, 0 divergences]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 73 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for details
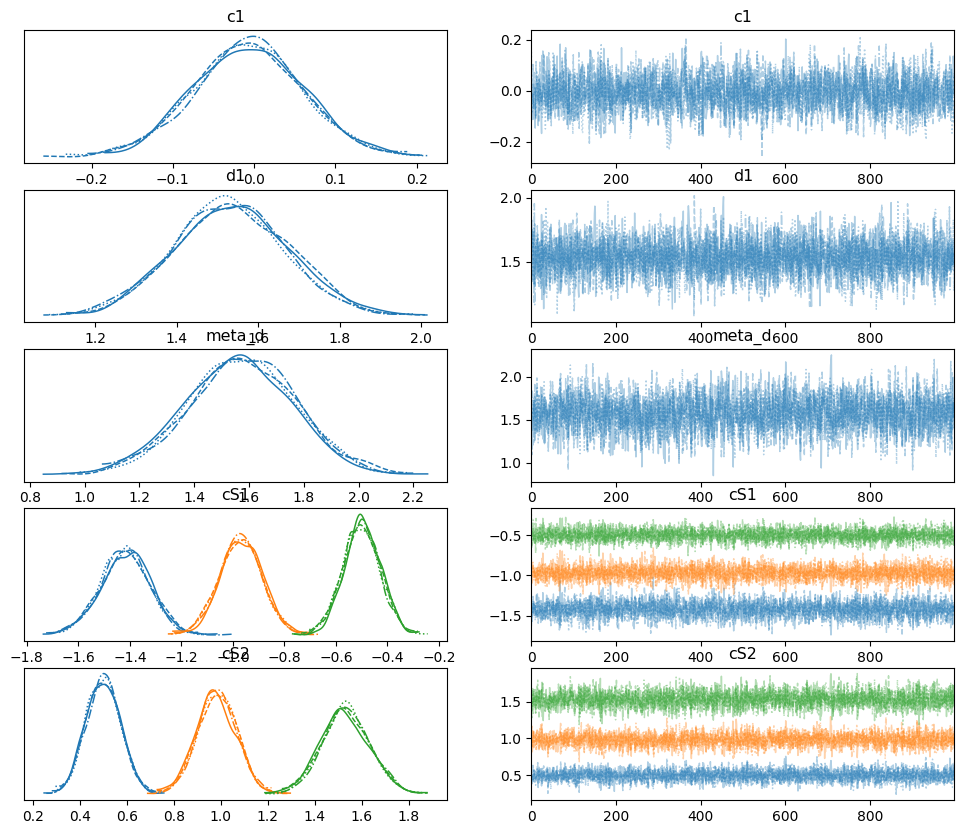
az.plot_trace(traces, var_names=["c1", "d1", "meta_d", "cS1", "cS2"]);
az.summary(traces, var_names=["c1", "d1", "meta_d", "cS1", "cS2"])
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| c1 | -0.007 | 0.067 | -0.137 | 0.114 | 0.001 | 0.001 | 2435.0 | 2688.0 | 1.0 |
| d1 | 1.532 | 0.139 | 1.279 | 1.801 | 0.002 | 0.001 | 4587.0 | 2882.0 | 1.0 |
| meta_d | 1.571 | 0.198 | 1.184 | 1.937 | 0.004 | 0.003 | 3027.0 | 3113.0 | 1.0 |
| cS1[0] | -1.418 | 0.095 | -1.595 | -1.238 | 0.002 | 0.001 | 3154.0 | 3135.0 | 1.0 |
| cS1[1] | -0.970 | 0.082 | -1.123 | -0.816 | 0.001 | 0.001 | 3284.0 | 3436.0 | 1.0 |
| cS1[2] | -0.500 | 0.071 | -0.626 | -0.360 | 0.001 | 0.001 | 3970.0 | 3069.0 | 1.0 |
| cS2[0] | 0.499 | 0.073 | 0.368 | 0.636 | 0.001 | 0.001 | 4407.0 | 3227.0 | 1.0 |
| cS2[1] | 0.983 | 0.084 | 0.822 | 1.135 | 0.001 | 0.001 | 3294.0 | 3579.0 | 1.0 |
| cS2[2] | 1.531 | 0.102 | 1.345 | 1.729 | 0.002 | 0.001 | 3102.0 | 3345.0 | 1.0 |
Watermark#
%load_ext watermark
%watermark -n -u -v -iv -w -p metadpy,pytensor,pymc
Last updated: Wed Nov 08 2023
Python implementation: CPython
Python version : 3.9.18
IPython version : 8.17.2
metadpy : 0.1.1
pytensor: 2.17.3
pymc : 5.9.1
sys : 3.9.18 (main, Aug 28 2023, 08:38:32)
[GCC 11.4.0]
numpy: 1.25.2
arviz: 0.16.1
Watermark: 2.4.3